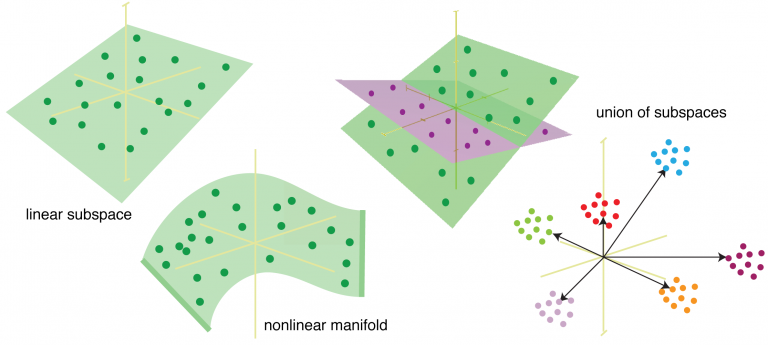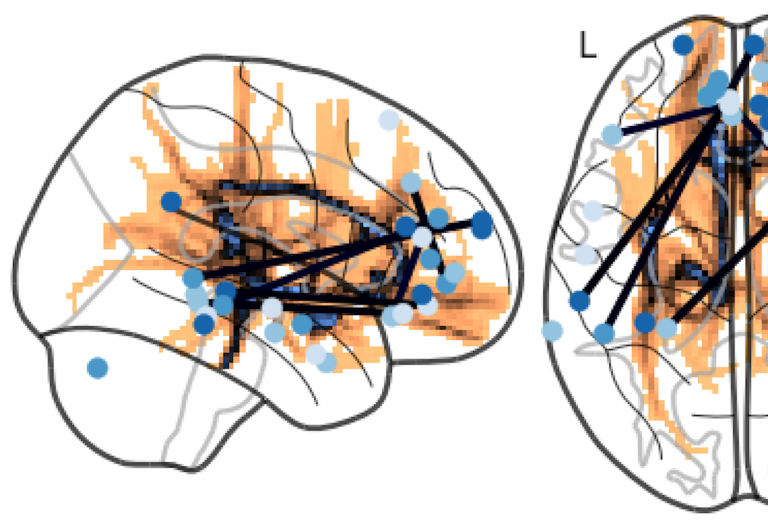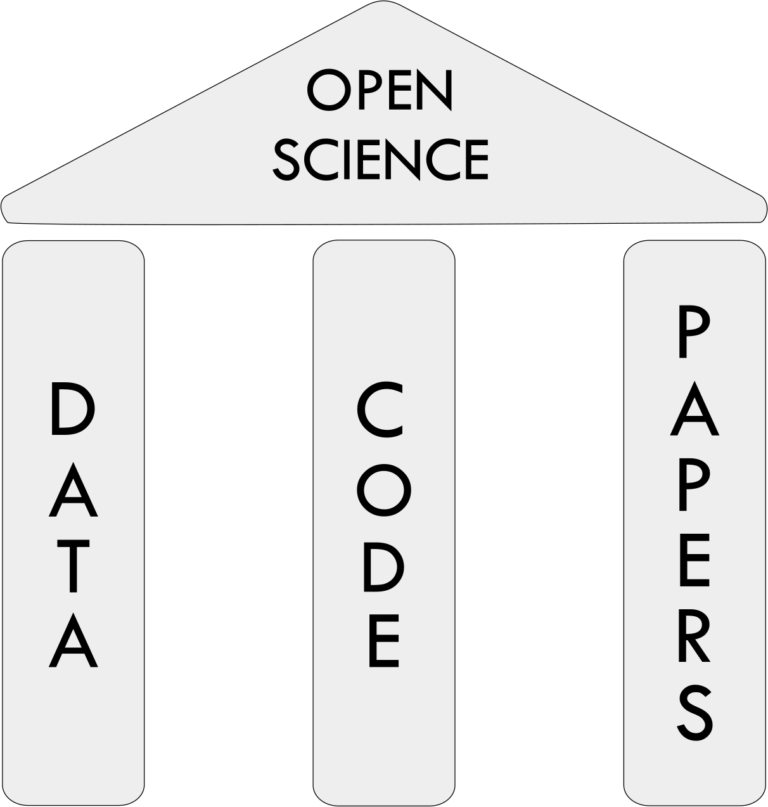
Brain hacking 101
60 minutes We’ll continue hacking all day thereafter.

Brain Imaging Data Structure (BIDS)
60 minutes
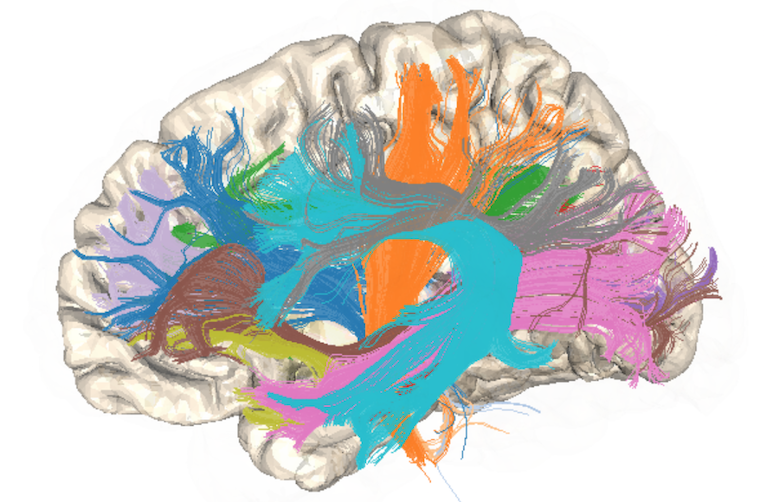
Breakout session
As a part of NeuroHackademy 2022, Melanie Ganz-Benjaminsen (University of Copenhagen), Taylor Salo (Florida International University), and Ariel Rokem (UW) provided a breakout session on BIDS and Liberty Hamilton (UT Austin) introduced receptive fields/encoding models.

Cloud computing for neuroimaging
60 minutes

Creating share-able Python libraries
90 minutes

CRN resources
The Center for Reproducible Neuroscience produces a variety of software resources (such as fmriprep and mriqc). In this tutorial, you will learn about some of them.

Cython and numba
Cython is a technology that allows us to easily bridge between python, and the underlying C representations. The main purpose of the library is to take code that is written in python, and, provided some additional amount of (mostly type) information, compile it to C, compile the C code, and bundle the C objects into […]

Data manipulation in Python (Numpy/Pandas)
90 minutes
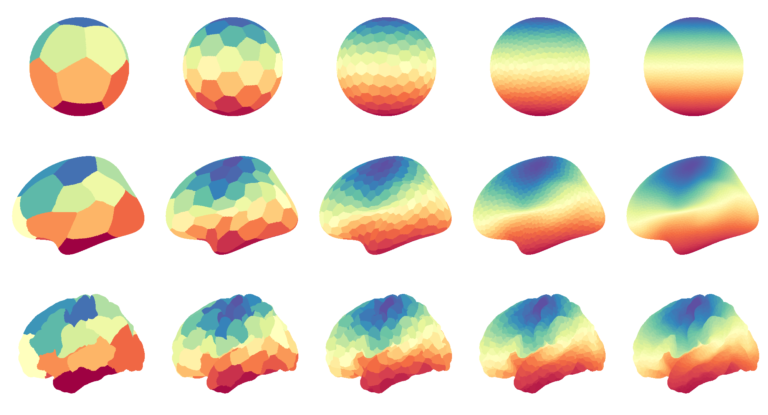
Data visualization in Python
90 minutes

Deep learning
This lecture will introduce PyTorch, a common machine learning library, and will demonstrate the use of PyTorch to develop a multi-layered neural network.

Docker
90 minutes
Docker for scientists
An overview of Docker and other containerization technologies: what containers are, why they’re useful, how to install them, and how to use them. Slides and materials available on GitHub: https://github.com/neurohackweek/docker-for-scientists.
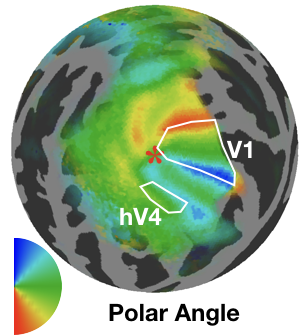
Functional brain parcellation
90 minutes

Git from Scratch
In this lecture we will learn why git is important in data science, how to use git via its command line interface, and how to configure git to interact with GitHub.

Git/Github
A tutorial on version control using git and GitHub.

High-performance Python
90 minutes

Introduction to Machine Learning
This presentation covers introductory machine learning concepts, including out-of-sample prediction, cross-validation, the variance/bias trade-off, and major classes of ML techniques. Application of these concepts to neuroimaging data is illustrated using the scikit-learn package for Python. 90 minutes
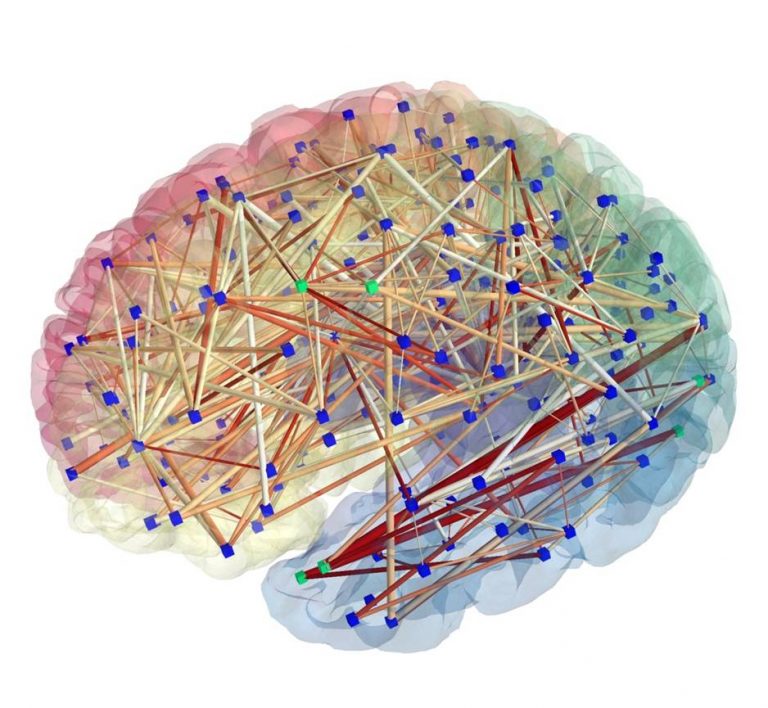
Introduction to NeuroHackademy
45 minutes

Introduction to Neurohackweek
Setting the stage, and explaining what will happen in the next couple of weeks.

Introduction to Neurohackweek
Introduces the concept of neurohackweek, the code of conduct, and the schedule for the week.

Introduction to programming in Python
A brief overview of the Python programming language, spanning both elementary concepts (variables and data types, control flow, etc.) as well as somewhat more advanced ones (object-oriented programming, argument unpacking, etc.). Includes a short overview of the Jupyter Notebook/Lab environment used for much of the instruction during this course.

Introduction to Python
A brief overview of the Python programming language, with an emphasis on tools relevant to data scientists. An interactive Jupyter Notebook (which also doubles as the slides) is available here.

Introduction to Python: Control Flow
This lecture will discuss control flow in Python: conditional statements, loops, and functions.

Introduction to Python: NumPy
This lecture will give a brief introduction to using NumPy, Python’s numerical library.

Introduction to Python: the Basics
This lecture will provide a ground-up introduction to Python. We will discuss Jupyter notebooks and how to use the JupyterHub, Python variables and types, and basic Python operations.

Introduction to the Geometry and Structure of the Human Brain
This talk will provide a hands-on introduction to the data structures, file formats, and tools used to represent and compute over anatomical and geometric properties of the human brain. Topics will include volumetric MRI data, cortical surface representations, and inter-subject comparison. 90 minutes
Machine learning for neuroimaging
90 minutes

Machine learning with scikit-learn
This session will cover the basics of Scikit-Learn, a popular package containing a collection of tools for machine learning written in Python. See more at http://scikit-learn.org. Outline Main Goal: To introduce the central concepts of machine learning, and how they can be applied in Python using the Scikit-learn Package. Definition of machine learning Data representation in scikit-learn […]
Meta-analysis and reproducibility
This talk will discuss neuroimaging meta-analysis as a tool for examining consensus and reproducibility, provide examples of different meta-analytic applications, and review related software and resources.
NiBabel: Neuroimaging data and file structures in Python
This talk will introduce neuroimaging data and file structures using the Python library NiBabel. It will describe the general classes of neuroimaging data researchers are likely to interact with – volumetric, surface, tractography, connectivity – and Python interfaces for loading, modifying and writing the data. There will be some discussion of particular file formats, memory […]

Nilearn
This talk will introduce the Python library nilearn, highlighting its use in bringing machine learning analyses to the neuroimaging domain. 90 minutes
Nipreps
90 minutes

Optimal transport (ADSI)
90 minutes

Optimization (ADSI)
This tutorial illustrates how many popular machine learning models can be formulated and solved through the lens of optimization. Through a combination of slides and exercises, we present intuition behind gradient descent, linear and logistic regression, regularization, and outlier detection. 90 minutes.

Panel discussion: Python ecosystem Q&A
An open discussion period for any/all questions regarding the Python language and the neuroimaging data science tools covered in this course. General questions regarding open-source software development (in any language), neuroinformatics, etc. are also welcome. 60 minutes.

Python packaging
In this tutorial, we will work through setting up a scientific Python package. This will provide an opinionated introduction to some of the ins and outs of Python packaging and package distribution.

Reproducibility in fMRI: What is the problem?
A review of barriers to reproducible neuroimaging research, and some potential solutions.
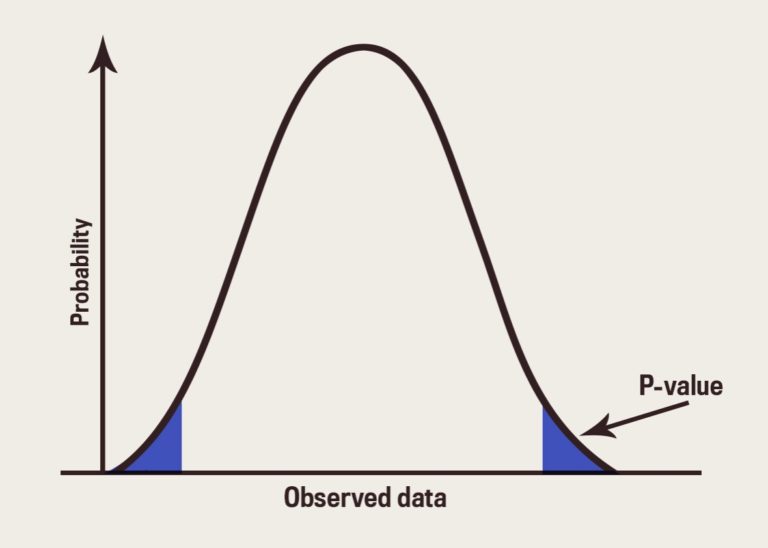
Reproducibility: what you need to know from epistemology to statistics
In this talk, I will first review the main causes impacting reproducibility and in particular the statistical aspects, in the context of classical statistics but also in prediction. Second, I will attempt to link these two frameworks to some classic epistemological concepts. 90 minutes

Science: open for all
How to practice and promote an inclusionary, welcoming open science. Slides can be found here.

Software testing
An introduction to software testing for scientific code. Materials available here; source code for all materials here.
The difference between prediction and explanation
This talk will overview the differences and overlap between machine learning and statistics as well as some important considerations when applying machine learning vs traditional statistics models to fMRI data. 90 minutes

Version control with git/GitHub
This talk will provide a hands-on introduction to git, the most popular version control software used today. We’ll showcase using the corresponding web portal GitHub, with an emphasis throughout on the importance of version control for collaboration and reproducibility in science. 90 minutes
Workflows/Nipype
This talk will introduce why workflow systems are essential for many scientific analyses. It will describe common concepts across workflow technologies used in biomedical research. It will then focus on the Nipype ecosystem as a set of tools to deliver robust and reproducible workflows for neuroimaging analysis. 90 minutes